Project Description
This project will analyze and visualize a tree constructed from rdp 16s database.
Input file
rdp_16s_v16_reads_50.fa: Random 50 reads from rdp 16s database
Tools need
reformat.sh: This will pick 50 random reads from rdp 16s database.
ETE Toolkit: Analysis and visualization of trees.
Commands
# Download 16s rdp database
wget https://www.drive5.com/sintax/rdp_16s_v16.fa.gz
# Select only 50 random reads
reformat.sh in=rdp_16s_v16.fa.gz out=rdp_16s_v16_reads_50.fa sample=50 overwrite=true
Align the reads and build the tree using ETE Toolkit
Clean the fasta file
# Remove " in the Fasta headers
sed -ie 's/\"//g' rdp_16s_v16_reads_50.fa
#Formant the Fasta headers and remove unwanted characters/strings
sed 's/;/,/' rdp_16s_v16_reads_50.fa | sed 's/tax=//'| sed 's/:/_/g' | sed 's/_[^~]*\,d/,d/' | sed 's,;,,' > rdp_16s_v16_reads_50_clean.fa
# Write headers into seperate file
grep ">" rdp_16s_v16_reads_50_clean.fa | sed 's/,/ /'| sed 's/>//' >fasta_headers.txt
# Remove any taxa information from header lines
sed -ie 's/,.*//' rdp_16s_v16_reads_50_clean.fa
For this purpose, basic workflow has been used. More
ete3 build -w standard_fasttree -n rdp_16s_v16_reads_50_clean.fa -o ./rdp_16s_tree --clearall
Display tree
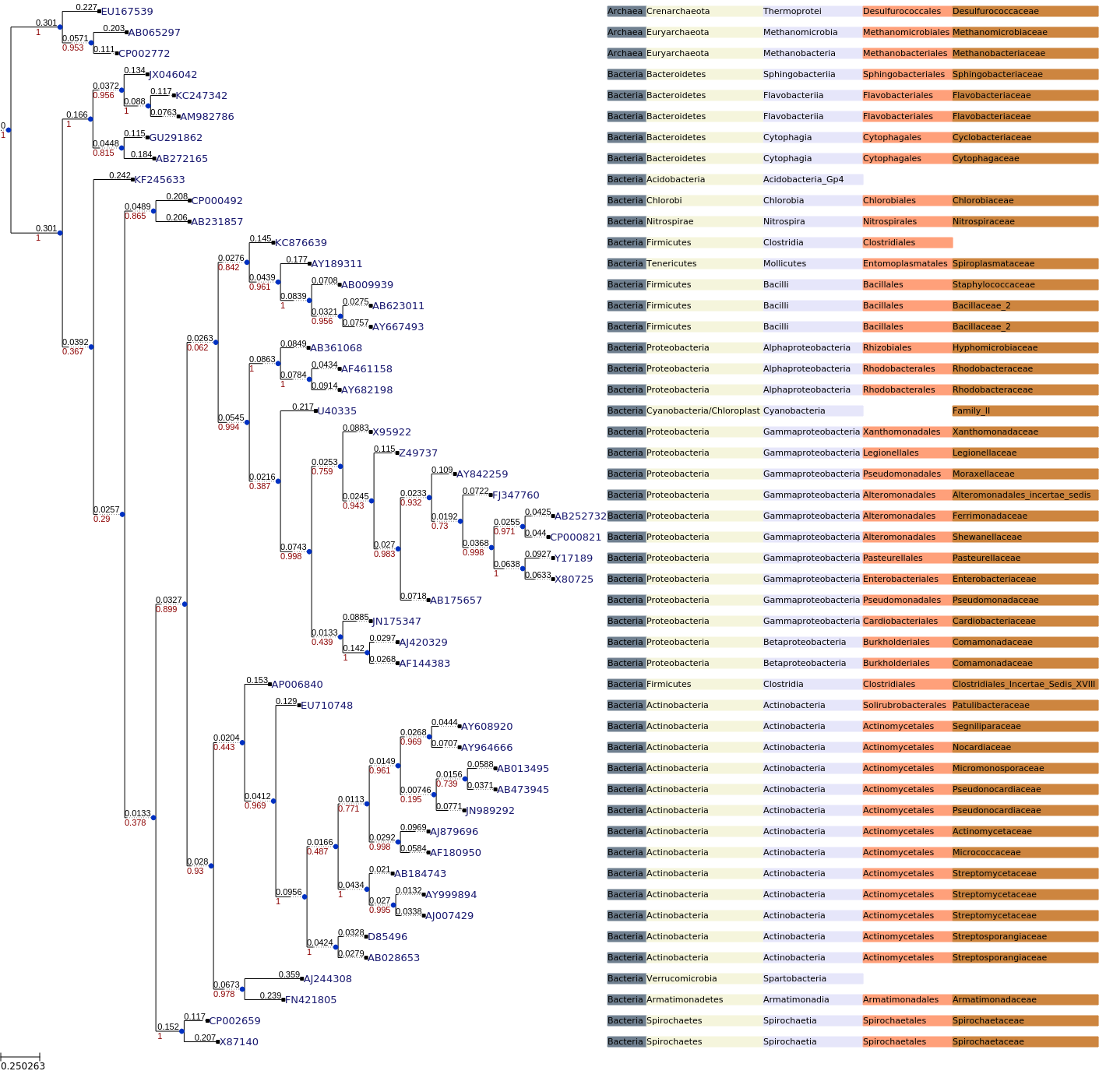
Following code will display the image of the Alignment.
from IPython.display import Image
Image(filename='rdp_16s_tree/clustalo_default-none-none-fasttree_full/rdp_16s_v16_reads_50_clean.fa.final_tree.png')
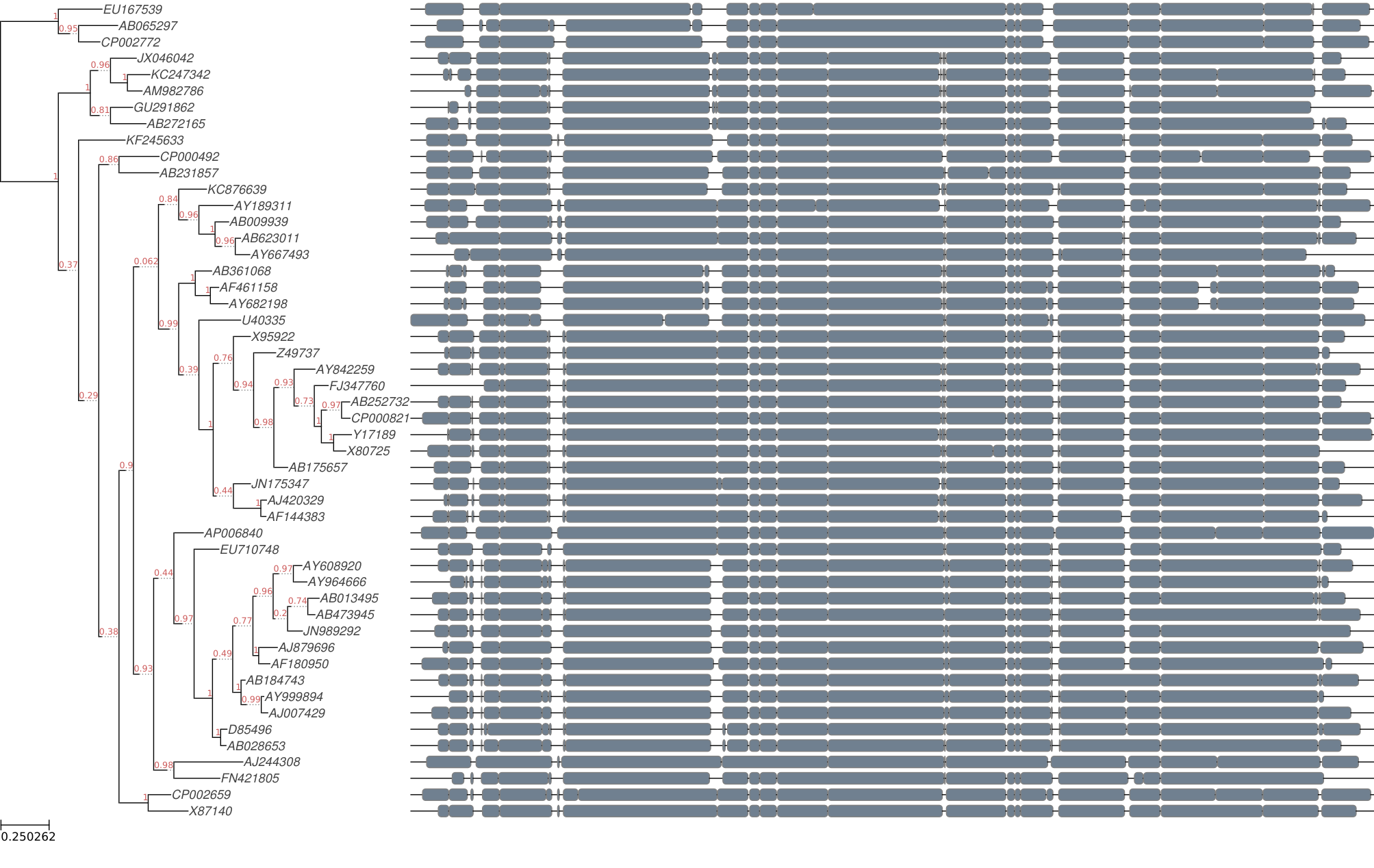
Display Tree with Alignment
Also, alignment can plot using ete3 module.
# Import Python libraries
from ete3 import PhyloTree, TreeStyle
# Import the tree using PhyloTree class
tree1 = PhyloTree("rdp_16s_tree/clustalo_default-none-none-fasttree_full/rdp_16s_v16_reads_50_clean.fa.final_tree.nw")
# Add alignment
tree1.link_to_alignment("rdp_16s_tree/clustalo_default-none-none-fasttree_full/rdp_16s_v16_reads_50_clean.fa.final_tree.used_alg.fa")
#tree1.render(file_name="rdp_alignment_tree.png", dpi=300 , w=1600, h=1000 )
# Render in jupyter notebook
tree1.render("%%inline", h=150, units="mm", dpi=100)

# Read the headers to annotate the tree
with open("./fasta_headers.txt", "r") as f:
headers=f.read().splitlines()
# Create the database of headers
header_dict={}
# Function to replace characters
def remove_level_identification_char(string):
# This will remove taxa level identification character
if string.startswith("d_"):
return {0:string.replace("d_", "")}
elif string.startswith("p_"):
return {1:string.replace("p_", "")}
elif string.startswith("c_"):
return {2:string.replace("c_", "")}
elif string.startswith("o_"):
return {3:string.replace("o_", "")}
elif string.startswith("f_"):
return {4:string.replace("f_", "")}
elif string.startswith("g_"):
return {5:string.replace("g_", "")}
for item in headers:
#temp dictionary to hold taxa levels
temp={}
node,taxa=item.split()
level_seperated = taxa.split(",")[:-1]
for i in level_seperated:
if temp:
temp.update(remove_level_identification_char(i))
else:
temp=remove_level_identification_char(i)
header_dict[node]=temp
The following colors are choosen to color each taxa level.
colour_palatte={0:"#708090", 1:"#F5F5DC", 2:"#E6E6FA", 3:"#FFA07A", 4:"#CD853F", 5:"#F08080"}
def custom_layout(node):
if node.is_leaf():
# Add an static face that handles the node name
faces.add_face_to_node(nameFace, node, column=0)
# Get the taxa levels corresponding to each mnode
lineage = header_dict[node.name]
# Plot each level with different colors
for key, level in lineage.items():
levelNameFace = faces.TextFace(level+" ", fsize=8, fgcolor= "Black")
levelNameFace.background.color = colour_palatte[key]
faces.add_face_to_node(levelNameFace, node, column=key, aligned=True)
from ete3 import Tree, faces, TreeStyle
# Nodes are colored green
nameFace = faces.AttrFace("name", fsize=10, fgcolor="#191970")
# TressStyle has been used to customize the image
ts = TreeStyle()
# Prvents node name showing twice
ts.show_leaf_name = False
# Shows the Tree branch length
ts.show_branch_length=True
# Boostrap values
ts.show_branch_support = True
# Uses Custome ayout created previously
ts.layout_fn = custom_layout
# plots the tree
tree1.render("%%inline", tree_style=ts)